Computational Biology Research Group

Computational Biology Research Group
We are committed to sharing the knowledge we acquired,
databases and algorithms we developed,
to support reproducing our work and to support efficiency in science.
The lab's research is driven by a conviction that internal protein structural districts/neighbourhoods, or, as we named them, Internal Protein Nanoenvironments (IPN), contain a significant core of information about their ultimate function. Such information content, fully describing corresponding nanoenvironments, is selectable in form of an ensemble of specific descriptors and corresponding values. The ensemble of physical-chemical and structural parameters is peculiarly less sensitive to localized variation of sequence encoding for that structure, causing limited structural promiscuity regarding underlining protein sequences, explaining why sequences may vary to a limited extent while resulting function remains unchanged. In conclusion: What we found is that for each nanoenvironment there is a specific ensemble of descriptors, making possible their cataloguing into a dictionary of IPNs. Also, the lab is continually employing leading initiatives to encourage and facilitate the use of “big data” in large-scale research across the scientific and technological disciplines. Finally, our lab focuses at the derivation of interactions in data that explain the observed correlations among IPNs and their resulting functions. That is the key element in structure/function relationship analysis and we identified that there is a significant potential for inferring such interactions in and for diverse internal protein districts.
1. Protein-Protein Interfaces (PPI)
2. Hot spots (HS)
3. Antibody-antigen interfaces (AA)
4. Protein-Ligand interfaces (PL)
5. Protein-DNA interfaces (PD)
6. Protein-Lipid membrane interfaces (PLM)
7. Protein channels
8. Secondary structure elements (SSE)
9. Cavities
10. Pockets
11. Catalytic site residues (CSR)
12. Allosteric sites (AS)
13. Exosites (ES)
14. Max distance reach for detection of AA Residue presence
This particular entry of the
Dictionary of Internal Protein Nanoenvironments (IPN)
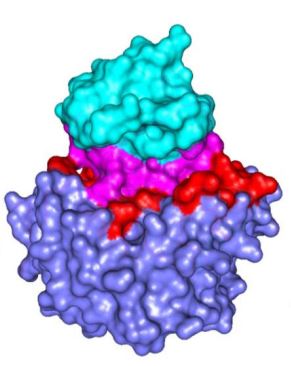
will examine biological structures with regard to Protein-Protein INTERFACES
and will offer to a user useful links such as the original paper
we published on this subject, its abstract, PhD theses elaborated
focusing on this specific issue.
Also, some graphical and tabular representations are selected for viewing of the key
biophysical, structural, biochemical and physical-chemical
descriptors of the NANOENVIRONMET described as Protein-Protein Interfaces.
Those are critical for understanding function of such
protein district and the whole protein.
The usefulness of this IPN Dictionary entry extends from agroindustrial and
pharmaceutical applications (model-based drug design for drugs acting at
protein interfaces, pharmacological activities) to some industrial
applications (understanding structural stability of protein polymers,
and in protein engineering).
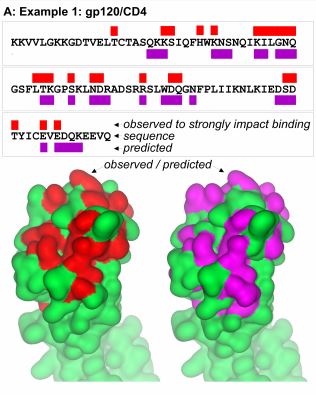
Through experiments of alanine scanning, it was shown that a small number of
residues belonging to protein interfaces contribute decisively to the binding
energy and so were called hot spots. Because of the importance of these residues
for protein-protein interactions many computational methods have been proposed
to predict the hot spots and thus complement the experimental procedure.
In this work, we developed methods to predict hot spots using support vector machines,
using at the input 186 structural descriptors extracted from the STING_DB and 112 new
descriptors proposed in this work. The methods proposed here showed superior performance
to methods of predicting hot spots best known from the literature, such as KFC, Minerva,
Rosetta and FOLDEF.
Here too, we will offer to a user useful links such as the PhD theses elaborated at the University of Campinas, SP, Brazil.
Viewing of the key biophysical,
structural, biochemical and physical-chemical descriptors for this NANOENVIRONMET is possible.
The usefulness of this IPN Dictionary entry extends from agroindustrial and pharmaceutical applications
(model-based drug design for drugs acting at protein interfaces, pharmacological activities) to some
industrial applications (understanding structural stability of protein polymers, and in protein engineering).
Antibodies are an important class of biological drugs, but with limitations, such as
inadequate pharmacokinetics, adverse immunogenicity and high production costs.
Synthetic peptides for the desired target represent an important alternative to antibodies.
To guide the design of these peptides it is necessary to fully understand the nanoenvironment that
governs specific interactions that guarantee precision and selectivity.
Here too, we present original paper, link to the software and plots with main descriptors
of this particular nanoenvironment.
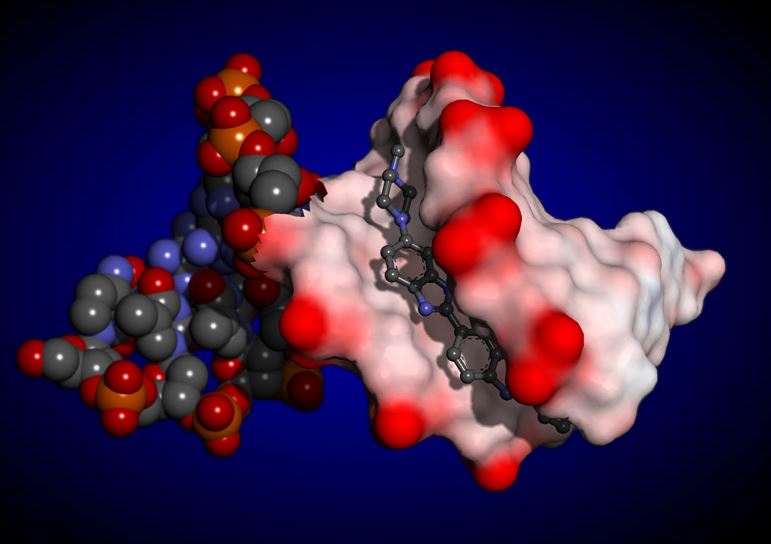
Having a better understanding of mechanisms for protein-ligand interactions is a key factor for
successful drug discovery projects. Here we focus on characterizing ligand-binding pockets which
are also designated as protein-ligand interaction sites: regions at the protein surface (and cavities)
where ligands bind and perform certain functional activity. We list here some of most important structural
and physicochemical features related to the molecular recognition process that involves protein from one
side and the corresponding ligand at the other side. Our hypothesis is that a more detailed
characterization of protein-ligand complexes in terms of describing ligand-binding sites by
its most relevant nanoenvironment descriptors (MRND can lead to improvements in identification
of such regions in general (for any protein), and in particular, a more precise ligand binding affinity prediction.
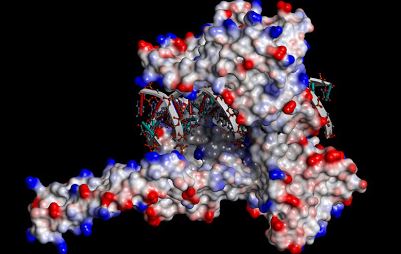
Protein-DNA interactions are key to control gene expression, transcription,
DNA repair and DNA packing among all living beings. Due to its importance,
several computational approaches that focus on the prediction of protein-DNA
interacting protein residues are available in the scientific literature. Yet,
only a fraction makes use of structural information and all of the available
methods rely on amino acids conservation. The methods described here about this
particular nanoenvironment are using statistical and machine learning approaches
having at the input some physicochemical and structural descriptors from Blue Star Sting
database that reflects the importance of each parameter in forming of the complex
between proteins and DNA molecules. The developed approach is important in order
to correctly assess protein function as well as identify important residues for
composing protein-DNA interfaces. Likewise, DNA component is also included in
terms of specific type of contacts established between two interacting macromolecules,
an extrimely valuble information for analysis of the protein-dna interface nanoenvironment.
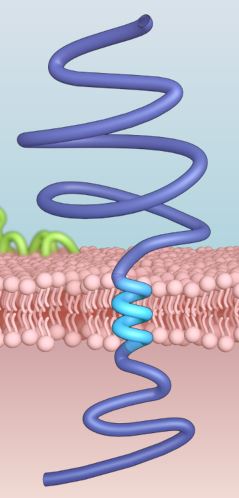
Here, we analyze the nanoenvironment of interfaces formed among lipid membranes and proteins.
Membrane proteins account for approximately one-third of the proteomes of all organisms and include receptors,
structural proteins and channels. We focus on two types of membrane proteins: integral membrane proteins which
are usually permanently anchored to the membrane,and so called peripheral membrane proteins, which are usually
temporarily attached to a lipid bilayer. The nanoenvironments of such interfaces represent potential
pharmacological targets of fundamental importance for a variety of diseases, with very important implications
for the design and discovery of new drugs or peptides modulating or inhibiting relevant interactions.
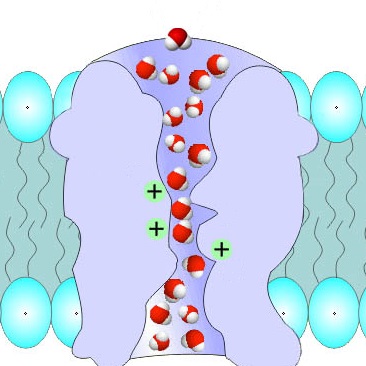
Protein channels are tunnel-like structures that traverse the surface or interior of proteins, facilitating the movement of ions,
molecules, or water across membranes or within the protein structure. These channels are typically lined with specific amino acid
residues that create a selective pathway, often tailored to the size, charge, or polarity of the transported substances.
Protein channels are essential in various biological processes, including signaling, metabolism, and transport. Their study provides
valuable insights into molecular mechanisms and aids in developing therapeutic agents targeting channel-associated functions or disorders.
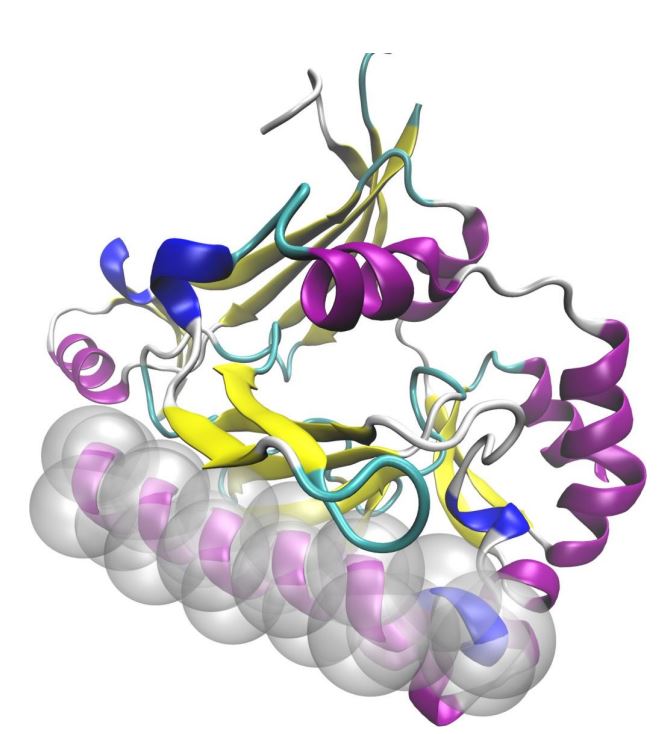
Protein secondary structure elements (PSSEs) such as α-helices, β-strands, and turns are the
basic building blocks of the tertiary protein structure. Our primary interest here is to
reveal the characteristics of the nanoenvironment formed by both PSSEs and their surrounding
amino acid residues (AARs), what might contribute to the general understanding of how
proteins fold. The characteristics of such nanoenvironments must be specific to each secondary structure
element, and we have set our goal here to gather the fullest possible description of the
α-helical nanoenvironment first, and then for β-strands and turns.
Here, published paper on this subject is presented as well as link to PhD theses with complete research data.
Graphical and tabular information abount nanoenvironments for α-helices, β-strands, and turns are offered for a user analysis.
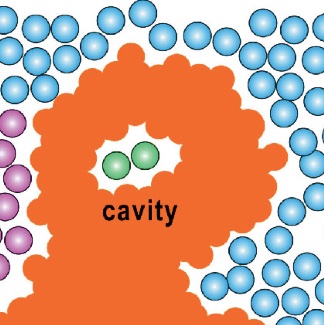
Protein cavities are enclosed or partially enclosed voids within the three-dimensional structure of proteins. These cavities can
play crucial roles in protein function, serving as sites for substrate binding, ion storage, catalysis, or even the appropriate "space buffer"
allowing for protein volume/shape changes, essential for function optimization. Their size, shape, and chemical environment are defined by
the arrangement of surrounding amino acid residues, often creating a specialized nanoenvironment that supports specific biochemical interactions.
Understanding and characterizing protein cavities is vital for elucidating protein mechanisms and for applications in drug design,
as they may provide opportunities to target otherwise challenging functional sites.
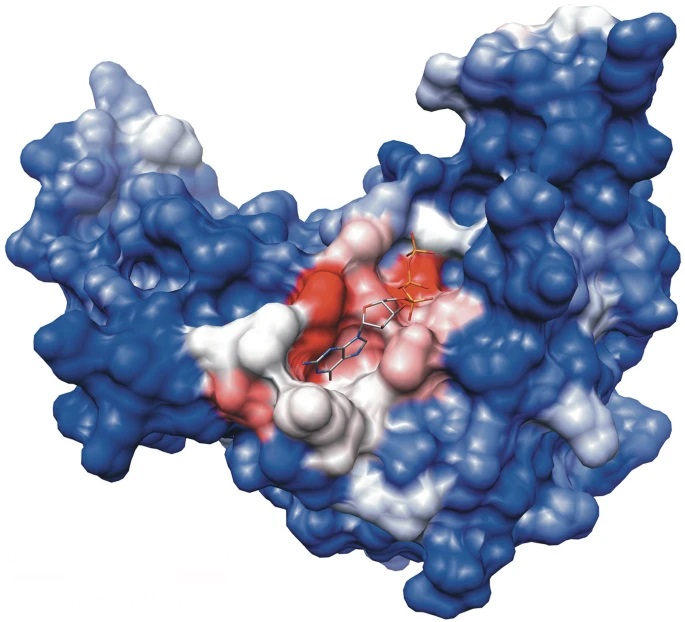
Protein surface pockets are structural indentations on the surface of proteins that often serve as key functional sites. These pockets can
host interactions with ligands, substrates, or other biomolecules, making them critical for enzymatic activity, rate control, molecular recognition,
and drug binding. Their shape, depth, and chemical properties are determined by the spatial arrangement of amino acids, and they can vary from shallow
grooves to deep cavities. Identifying and characterizing these pockets is essential in structural biology and drug design, as they offer insights into
protein function and potential therapeutic targets.
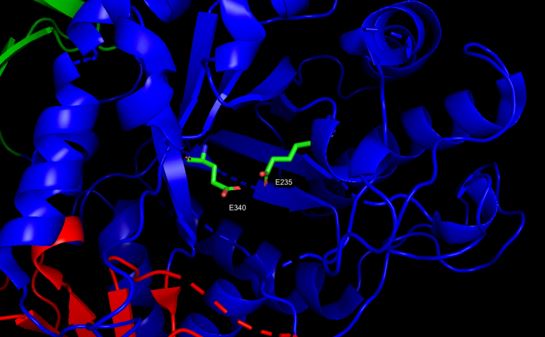
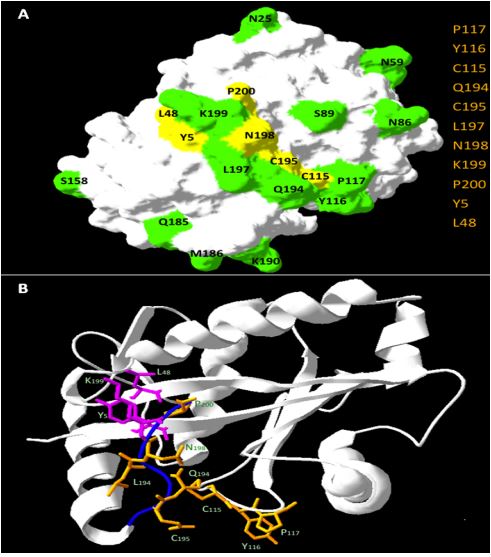
The function of enzymes is determined by specific residues , called catalytic amino
acids residues(CSR). The protein function is maintained for eons of selective
pressure which preserves in its structure many physical-chemical and structural patterns.
Frequently, enzymes from distinct organisms exert exactly the same biological function
due to preservation of similar catalytic amino acid residues, even with evident
low sequence similarity at the level of whole proteins.
The majority of catalytic amino acid residues prediction methods use sequence
conservation features to provide classification. Seeking to understand these
conserved patterns in enzyme structures, that even after eons of evolution
perform the same biological function, the present work searches to identify
which protein structural descriptors (available in Blue Star STING platform)
are capable of discriminating the amino acid catalytic residues from non-catalytic
residues by means of their nanoenvironment properties.
Here too, we will offer to a user useful links such as the PhD theses elaborated at the University of Campinas, SP, Brazil.
Viewing of the key biophysical,
structural, biochemical and physical-chemical descriptors for CSR NANOENVIRONMET is possible.

High-resolution structural data has been instrumental in characterizing and defining the stereochemical parameters that promote and define the binding of peptides, protein inhibitors and substrates at the active sites of enzymes to the point that we now have a very comprehensive understanding of specific interactions and catalytic mechanisms which facilitates the design and development of highly selective synthetic inhibitors and drugs. On a more subtle level, protein surfaces often bristle with secondary binding sites which serve key roles in regulating and modulating diverse activities ranging from gene expression, conformational stabilization, transport, inhibition and, by extension, the modulation and exhibition of distinct functions or moonlighting by the same enzyme in response to changes in physicochemical conditions is less well understood. Here we are compiling key descriptors for allosteric forming residues (AFRs) so we may gain more insight on how the site, function and how they regulate the main protein function and are being regulated themselves.
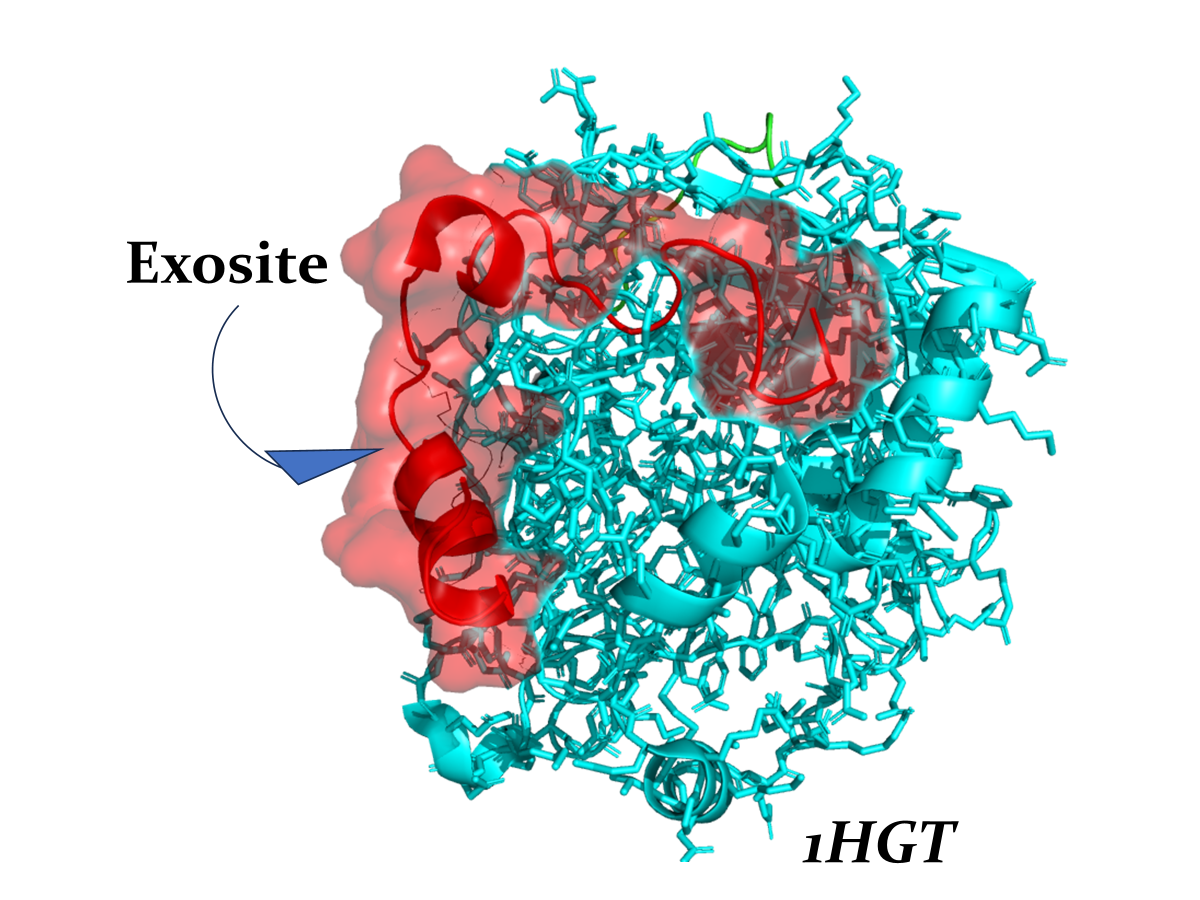
Exosites are secondary binding regions located on the protein surface that are distinct from both the active site and traditional allosteric sites. Unlike allosteric sites, exosites primarily facilitate protein-protein or protein-ligand interactions without directly influencing catalytic activity. Instead, they serve critical roles in fine-tuning protein function by mediating conformational stability, substrate recognition, or facilitating regulatory networks.
Exosites have been implicated in moonlighting functions, where a single protein exhibits distinct activities depending on physicochemical conditions or interaction partners. Here we study this nanoenvironment including its structural and dynamic properties, providing new opportunities for targeted therapeutic intervention and the development of biomolecular tools for probing this protein function.
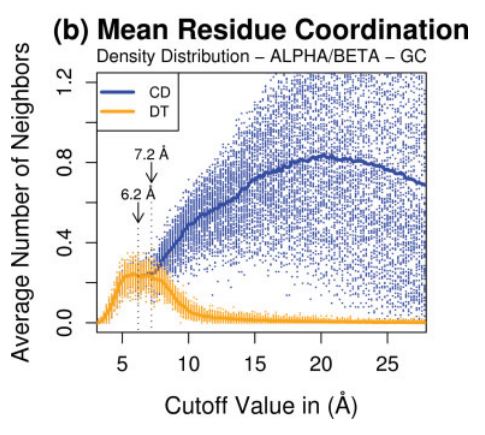
This particular entry in our dictionary of IPNs is not strictly speaking an environment. Rather, it is an feature of all
nanoenvironments within a protein structure, which determines the extension or reach of presence of any amino acid residue
"felt" across distance. For that, our work on atom coordination is presented as a useful tool for internal protein nanoenvironment
understanding, in particular, establishment of contacts and "presence" among AAR - a very important descriptor in STING RDB.
Atom and residue contacts have been used in a wide range
range of studies involving proteins and other biomolecules. Its correct and precise assignment comprise
the touchstone of the most important structural analysis
algorithms, which should be able to perform: packing
calculations, functional similarities, evolutionary
relationships, topological classifications, structural
alignments, structural assessment, protein structure
prediction, threading experiments, network contact analysis, empirical potentials,
thermodynamic stability previews, folding inferences, protein–protein and protein–ligand interactions,
and so forth.
Here we will focus our attention on some methods that underlay contact characterizations in
most of these applications.